Command-line Based Practical RNA-Seq Data Analysis With Linux & R (Subscription)
About Course
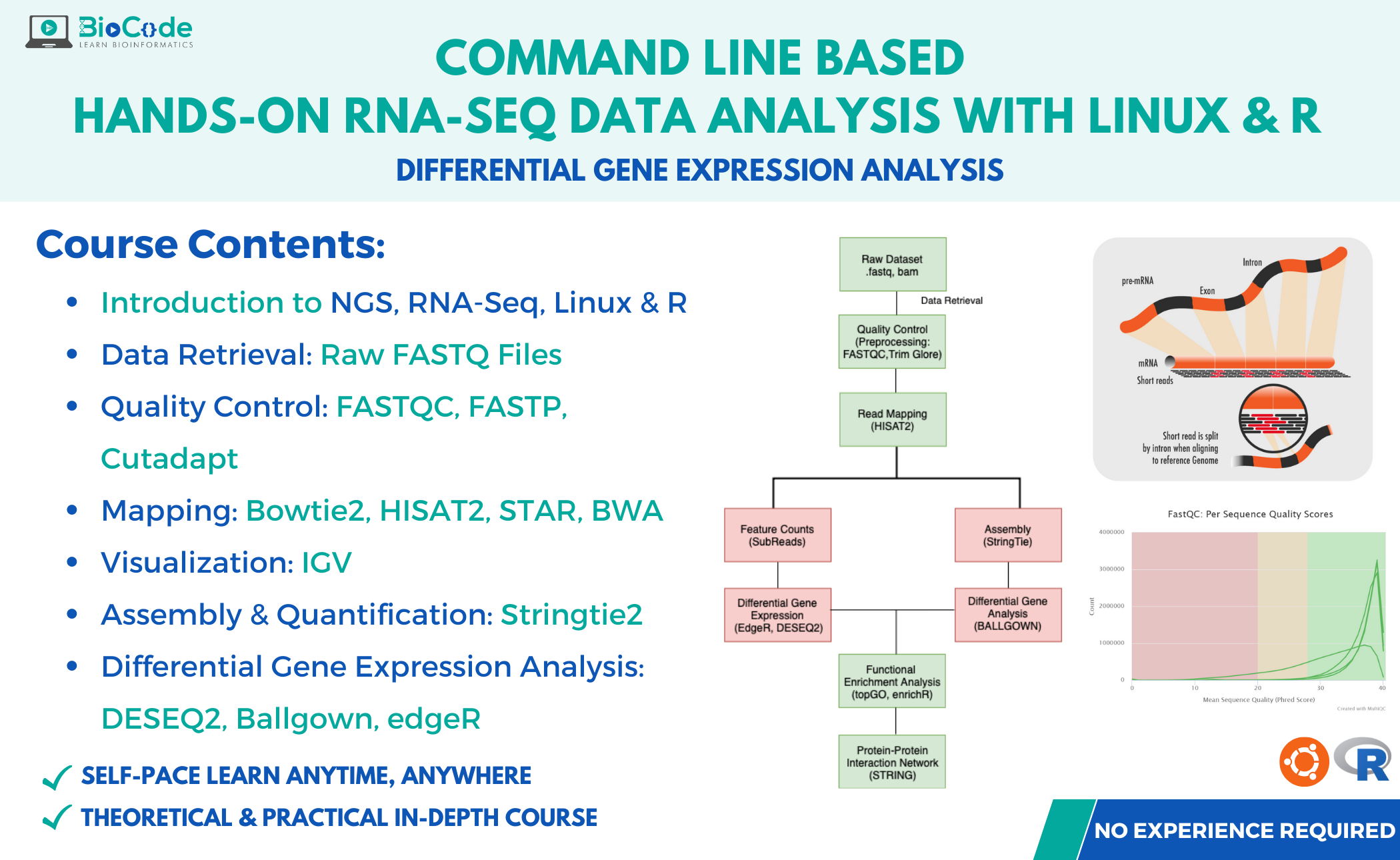
What Will You Learn?
- Introduction to RNA-Seq, NGS
- In-depth Knowledge of R & Linux
- Command-line tools for RNA-Seq
- Quality Control and Trimming
- Mapping & Evaluation of Alignment
- Discovering DEGs with DESeq2, edgeR and Ballgown
- Functional Analysis with Gene Ontology & KEGG Pathways
Course Content
In-Depth Introduction to RNA-Seq
-
In-depth Introduction to RNA-Seq Data Analysis, Linux Computing and R
44:08 -
Introduction to Linux for Bioinformatics
22:32 -
Introduction to ArrayExpress – Getting Started With MicroArray Analysis
09:56
Genomic Databases for Raw Data
-
BioProject
06:40 -
BioSystems
04:16 -
BioSample
02:56 -
Introduction to Gene Expression Omnibus Database
09:16 -
Gene Expression Omnibus – Platforms
05:42 -
Gene Expression Omnibus – Samples
04:16 -
Gene Expression Omnibus – Series
04:01 -
Gene Expression Omnibus – Datasets
04:45 -
Gene Expression Omnibus – Profiles – How to Search for Gene Expression Profiles
05:55 -
NCBI Genomes & NCBI Assembly
36:14 -
Genome Reference Consortium
07:48 -
Introducton to Phytozome
09:39 -
Download an Entire Plant Genome & Proteome
26:41 -
Introduction to UCSC Genome Browser
13:40 -
How to retrieve an entire genome & Retrieval of SARS-CoV-2 Viral Genome
09:41 -
Introduction to ENSEMBL
07:50 -
Genome Assembly Retrieval and Analysis
10:24
File Formats
-
BED
04:27 -
GTF/GFF
11:07 -
SAM
09:07 -
FASTQ
18:02
Introduction to Linux for RNA-Seq
-
Introduction to Linux for Bioinformatics
22:32 -
PWD – Print Working Directory
01:26 -
MKDIR – Making Directories
08:13 -
CD – Changing Directories
05:03 -
MV – Moving Files, Directories and Data
05:11 -
RM – Deleting Files and Directories
01:24 -
Which & Whereis – Find Programs You Installed
03:43 -
Find – Finding User Created Files
03:39 -
LS – Listing Files and Directories on Linux
06:46 -
Cat – Visualization and Inspection of Text Data
03:56 -
Head – Reading Specified Number of Lines from Top
03:50 -
Tail- Reading Specified Number of Lines from Bottom
02:23 -
Touch – Modifying File Statistics and Creating Files
07:04 -
Stat – Statistics of File & Directories
02:43 -
Wget – Retrieval of Genome Assemblies
06:48 -
Curl – Retrieval of Bioinformatics Files
02:25 -
Vim – Create and Edit Text Files
05:59 -
Diff – Find Sequence Differences in Files
02:35
RNA-Seq Data Analysis Pipeline (Theoretical & Practical)
-
How to Retrieve Raw Reads for RNA-Seq Data Analysis
03:26 -
Retrieval of Data Required for this Course
08:45 -
In-depth Theory: Quality Control
19:37 -
Practical: Preprocess Raw Reads Using FASTQC, FASTP, Trimmomatic & Cutadapt
31:54 -
In-depth Theory: Mapping & Transcriptome Assembly
48:22 -
Practical: Mapping Preprocessed Reads Against a Reference Genome Using HISAT2, TOPHAT2, BOWTIE2, STAR
31:51 -
Practical: Transcriptome Assembly Using StringTie
54:31 -
Mapping Visualization & Evaluation
36:37 -
In-depth Theory: Gene Quantification, Expression Estimation & Differential Gene Expression
18:28 -
In-depth Theory: DeSeq2, Ballgown, edgeR
32:22 -
Practical: Differential Gene Expression Analysis Using Ballgown, edgeR, DESEQ2 in R
34:39 -
Theory: Functinal Enrichment Analysis of the DEGs
18:39 -
Practial: Gene Ontology Analysis
28:02 -
Practical: Pathways Analysis Using KEGG, PANTHER, Reactome
12:41 -
Practical: Protein-Protein Interaction Using STRING
13:17
Evaluation
-
Exercise
Earn a certificate
Add this certificate to your resume to demonstrate your skills & increase your chances of getting noticed.

Student Ratings & Reviews

No Review Yet
Blue Area, Islamabad, Pakistan
Office: 4 Mann Island, Liverpool, Merseyside, United Kingdom
Tel: +44 7756207003
Email: support@biocode.org.uk
